近期发表文章 (#Co-corresponding author, *Co-first author)
1. Xie, Y. *, Zhang, T. *, Yang, M. *, Lyu, H., Zou, Y., Sun, Y., Xiao, J., Lian, W., Tao, J., Han, H., and Xu, C. (2025). Engineering crop flower morphology facilitates robotization of cross-pollination and speed breeding. Cell S0092-8674(25)00840-2.
2. Lou, H.*, Li, S.*, Shi, Z., Zou, Y., Zhang, Y., Huang, X., Yang, D., Yang, Y., Li, Z., and Xu, C. (2025). Engineering source-sink relations by prime editing confers heat-stress resilience in tomato and rice. Cell 188, 530-549.
3. Huang, X.*, Xiao, N.*, Xie, Y., and Xu, C. (2025). ROS burst prolongs transcriptional condensation to slow shoot apical meristem maturation and achieve heat-stress resilience in tomato. Developmental Cell 60(15), 2032–2045.
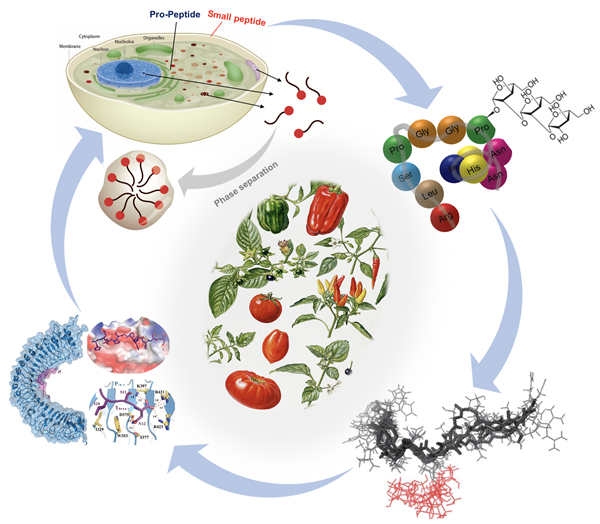
4. Yu, Y., Chu, J., Dong, S., Song, W. and Xu, C. Sugar codes for plant fitness: arabinosylation in small peptide signaling. Trends in Plant Science S1360-1385(25)00219-5
5. Yu, X.*, Li, Z.*, Yang, Y.*, Li, S., Lu, Y., Li, Y., Zhang, X., Chen, F., and Xu, C. (2025). Harnessing Green Revolution genes to optimize tomato production efficiency for vertical farming. Journal of integrative plant biology 67(9), 2446–2460.
6. Huang, X.*, Xiao, N.*, Zou, Y., Xie, Y., Tang, L., Zhang, Y., Yu, Y., Li, Y., and Xu, C. (2022). Heterotypic transcriptional condensates formed by prion-like paralogous proteins canalize flowering transition in tomato. Genome Biology 23, 78.
7. Huang, X.*, Chen, S.*, Li, W., Tang, L., Zhang, Y., Yang, N., Zou, Y., Zhai, X., Xiao, N., Liu, W., Li, P.#, and Xu, C.# (2021). ROS regulated reversible protein phase separation synchronizes plant flowering. Nature Chemical Biology 17, 549-557.
8. Xie, Y., Zhang, T., Huang, X., and Xu, C. (2022). A two-in-one breeding strategy boosts rapid utilization of wild species and elite cultivars. Plant Biotechnology Journal 20, 800-802.
9. Kwon, C.-T., Tang, L., Wang, X., Gentile, I., Hendelman, A., Robitaille, G., Van Eck, J., Xu, C.#, and Lippman, Z.#. (2022). Dynamic evolution of small signalling peptide compensation in plant stem cell control. Nature plants 8, 346-355.
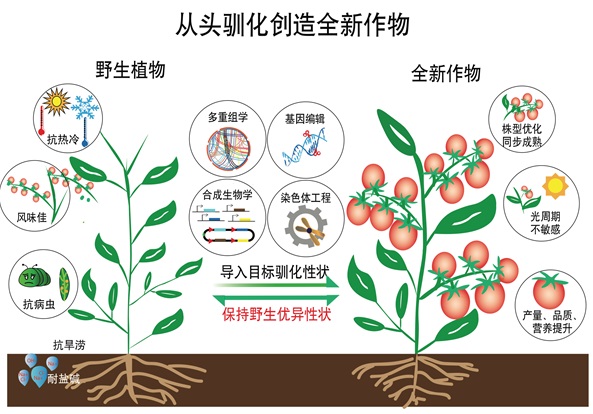
10. Li, T.*, Yang, X.*, Yu, Y.*, Si, X., Zhai, X., Zhang, H., Dong, W., Gao, C.#, and Xu, C.#. (2018). Domestication of wild tomato is accelerated by genome editing. Nature Biotechnology 36, 1160-1163.
11. Yu, Y, Xu, C. (2025). Decoding the cellular symphony of leaf senescence and nutrient allocation. Seed Biology DOI:10.48130/seedbio-0025-0010.
12. Yang, D., and Xu, C. (2025). When lettuce bolts: natural selection vs artificial selection and beyond. New Phytologist 246(3), 818–820.
13. Huang, X., Yang, Y., and Xu, C. (2025). Biomolecular condensation programs floral transition to orchestrate flowering time and inflorescence architecture. New Phytologist 245, 88-94.
14. Huang, X., and Xu, C. (2025). Reviving the charm of a century-old classic theory: Engineering source-sink relations to breed climate-smart crops. The Innovation Life 3, 100125.
15. Liu, N.*, Zou, Y.*, Jiang, Z.*, Tu, L., Wu, X., Li, D., Wang, J., Huang, L.#, Xu, C.#, and Gao, W#. (2024). Multiomics driven identification of glycosyltransferases in flavonoid glycoside biosynthesis in safflower. Horticultural Plant Journal. https://doi.org
/10.1016/j.hpj.2024.01.016.
16. Chen, S.*, Zou, Y.*, Tong, X., and Xu, C. (2024). A tomato NBS-LRR gene Mi-9 confers heat-stable resistance to root-knot nematodes. Journal of Integrative Agriculture https://doi.org/10.1016/j.jia.2024.07.017.
17. Yang, D., Wang, Z., Huang, X., and Xu, C. (2023). Molecular regulation of tomato male reproductive development. aBIOTECH 4, 72-82.
18. Wu, Q.#, Schmidt, W.#, Aalen, R.B.#, Xu, C.#, and Takahashi, F#. (2022). Editorial: Peptide signaling in plants. Frontiers in Plant Science 13, 843918
19. Xu, C. (2021). From 0 to 1: de novo domestication of allotetraploid wild rice to create a new crop. Hereditas (Beijing) 43, 199-202.
20. Tu, L.*, Su, P.*, Zhang, Z.*, Gao, L., Wang, J., Hu, T., Zhou, J., Zhang, Y., Zhao, Y., Liu, Y., Song, Y., Tong, Y., Lu, Y., Yang, J., Xu, C., Jia, M., Peters, R.J., Huang, L.#, and Gao, W#. (2020). Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nature communications 11, 971.
21. Zhao, H., Qin, Y., Xiao, Z., Li, Q., Yang, N., Pan, Z., Gong, D., Sun, Q., Yang, F., Zhang, Z., Wu, Y., Xu, C., Qiu, F. (2020). Loss of function of an RNA polymerase III subunit leads to impaired maize kernel development. Plant Physiology 184, 359-373.
22. Rodriguez-Leal, D.*, Xu, C.*, Kwon, C.-T., Soyars, C., Demesa-Arevalo, E., Man, J., Liu, L., Lemmon, Z.H., Jones, D.S., Van Eck, J., Jackson, D.P.#, Bartlett, M.E.#, Nimchuk, Z.L.#, and Lippman, Z.B.# (2019). Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nature Genetics 51, 786-792.
23. Huang, X., Tang, L., Yu, Y., Dalrymple, J., Lippman, Z.B.#, and Xu, C.# (2018). Control of flowering and inflorescence architecture in tomato by synergistic interactions between ALOG transcription factors. Journal of Genetics and Genomics 45, 557-560.
22. Zhang, N.*, Yu, H.*, Yu, H.*, Cai, Y., Huang, L., Xu, C., Xiong, G., Meng, X., Wang, J., Chen, H., Liu, G., Jing, Y., Yuan, Y., Liang, Y., Li, S., Smith, S.M., Li, J., and Wang, Y. (2018). A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. The Plant cell 30, 1461-1475.
23. Xu, C., Park, S.J., Van Eck, J., and Lippman, Z.B. (2016). Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes & Development 30, 2048-2061.
24. Xu, C.*, Liberatore, K.L.*, MacAlister, C.A., Huang, Z., Chu, Y.-H., Jiang, K., Brooks, C., Ogawa-Ohnishi, M., Xiong, G., Pauly, M., Van Eck, J., Matsubayashi, Y., van der Knaap, E., and Lippman, Z.B. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nature Genetics 47, 784-792.
25. Xu, C.*, Wang, Y.*, Yu, Y.*, Duan, J., Liao, Z., Xiong, G., Meng, X., Liu, G., Qian, Q.#, and Li, J.# (2012). Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nature communications 3, 750.